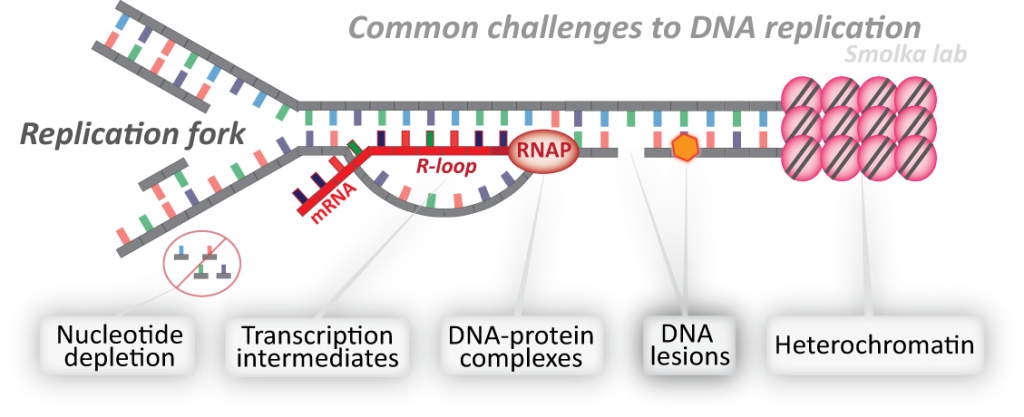
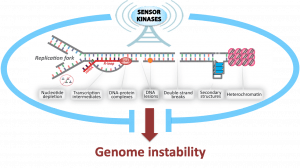
The integrity of our genome is especially at risk while it is being replicated. During DNA replication, the DNA must be “unzipped”, giving rise to structures known as replication forks. While traversing the genome, replication forks often encounter obstacles to their progression, including DNA lesions, hard-to-replicate sequences, transcription intermediates, or protein-DNA complexes. These encounters are potential sources of DNA breaks, chromosomal rearrangements and aneuploidy, all of which are hallmarks and drivers of cancer. Proper control of replication fork progression is therefore essential for genome integrity and cancer avoidance. Paradoxically, numerous anti-cancer agents such as topoisomerase inhibitors, DNA crosslinkers and DNA alkylators, kill cancer cells by impairing the regulated progression of the replication machinery and inducing replication stress. Our laboratory studies how cells sense, signal and prevent DNA replication stress to ensure faithful genome replication. We seek a deep mechanistic and integrated understanding of the replicative stress response and its implications for tumorigenesis and improved cancer treatment.
Signaling networks required for genome integrity, and their connections to oncogenesis, therapy and reproduction. We are investigating how the actions of kinases are translated into a coordinated cellular response that ensures proper maintenance and replication of the genome. Our recent work in yeast and mammalian systems uncovered how phosphorylation events mediated by DNA damage signaling kinases form a “code” of combinatorial protein interactions leading to a range of distinct functional outputs in the control of DNA replication, cell cycle and DNA repair. Recent work is also beginning to bridge our fundamental work toward potential clinical implications, including studies on the rational use of inhibitors of DNA damage kinases to manipulate DNA repair systems in cancer cells. We are also mapping the action of DNA damage signaling kinases in mammalian meiosis to define their roles in preventing infertility.
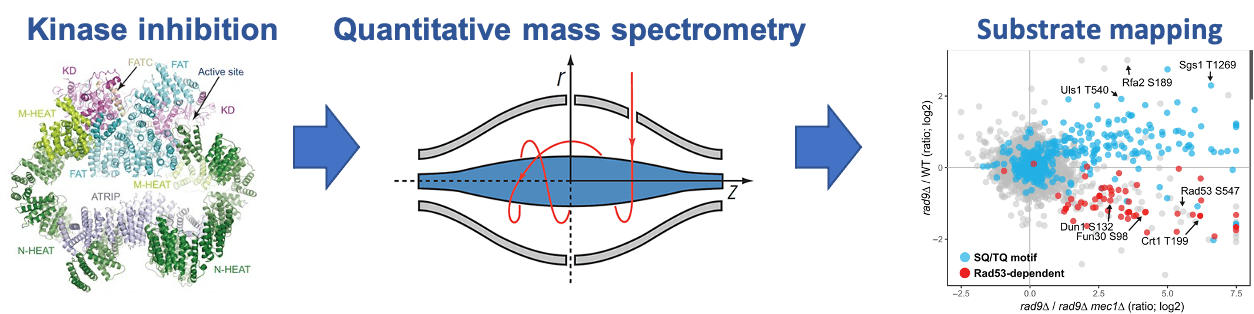
Phosphoproteomics. Reversible protein phosphorylation is widely used by cells as a signaling mechanism. Understanding the molecular basis of kinase action and function requires knowledge of the kinase substrates, as well as comprehensive characterization of the dynamics and role of the phosphorylation events. Because many kinases are active in a cell and thousands of proteins are phosphorylated, the study of phosphorylation-mediated signaling pathways is challenging and powerful technologies are needed. We have developed and applied quantitative mass spectrometry technologies for the phosphorylation analysis of protein complexes and for global screens of in vivo kinase substrates. We are now expanding the use of these technologies to quantitatively characterize signaling dynamics and regulation at a proteome-wide scale in a range of cellular systems and tissues.
We are grateful for the funding provided by: